Electrostatics and ions in biology
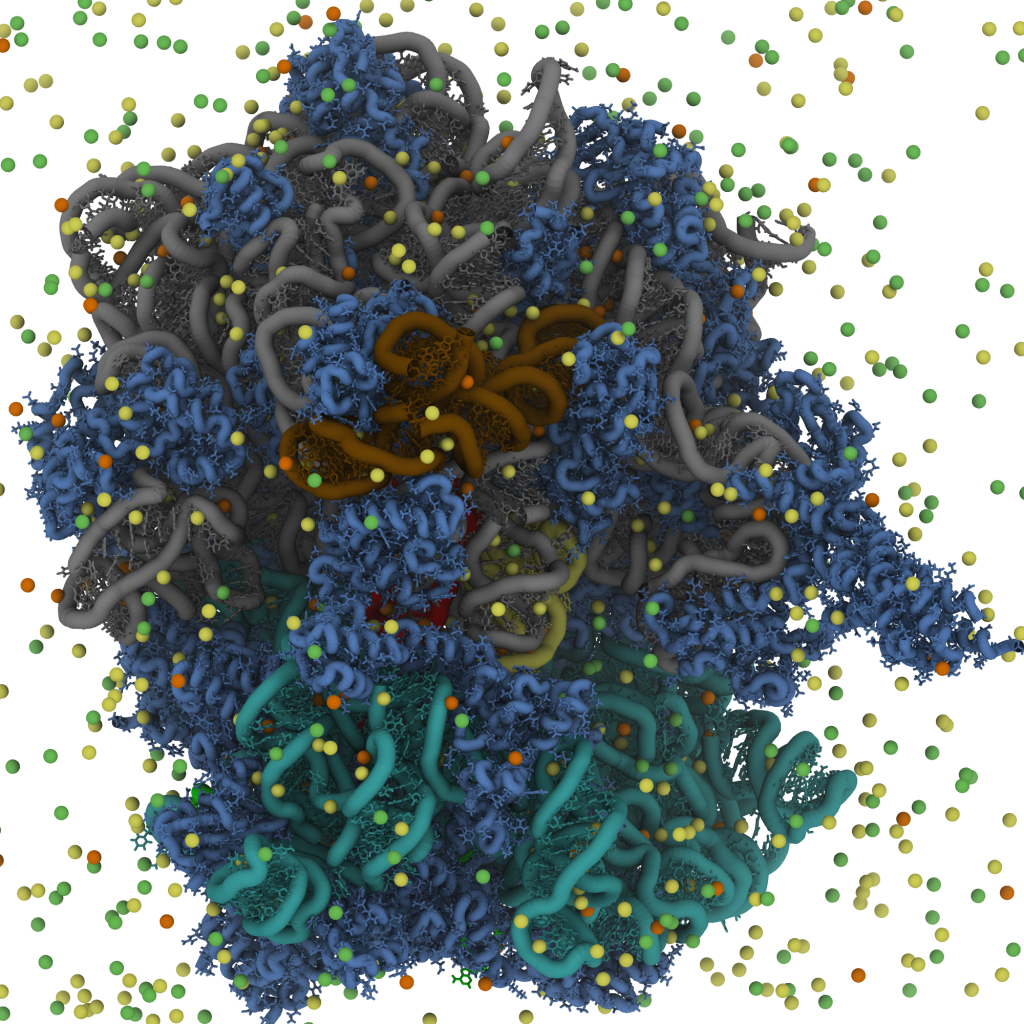
 While it is well known that precise ionic conditions are required for stability and function of biomolecules, in particular RNA, quantifying their specific roles has been particularly challenging. One reason for this is that they have many modes by which they can impart stability. When they bind stably, they may be directly probed by structural biology methods (X-ray, cryo-EM, NMR). Even though these strongly-bound ions are very important for biological function, a larger number of ions associate more transiently. These transient ions can be associated with short-range (unidentate ‘chelated’), intermediate-range (‘diffuse’) and long-range (‘bulk’) interactions.
While it is well known that precise ionic conditions are required for stability and function of biomolecules, in particular RNA, quantifying their specific roles has been particularly challenging. One reason for this is that they have many modes by which they can impart stability. When they bind stably, they may be directly probed by structural biology methods (X-ray, cryo-EM, NMR). Even though these strongly-bound ions are very important for biological function, a larger number of ions associate more transiently. These transient ions can be associated with short-range (unidentate ‘chelated’), intermediate-range (‘diffuse’) and long-range (‘bulk’) interactions.
In the study of ions, we recently developed an explicit-ion model that includes diffuse Mg2+, Cl- and K+ ions (Wang et al, JACS, 2022). Using this model, we were able to perform initial simulations of tRNA accommodation on the ribosome. This allowed us to quantify the concentration dependence of the free-energy landscape of the ribosome, while also identifying specific ion-mediated interactions that control the kinetics.
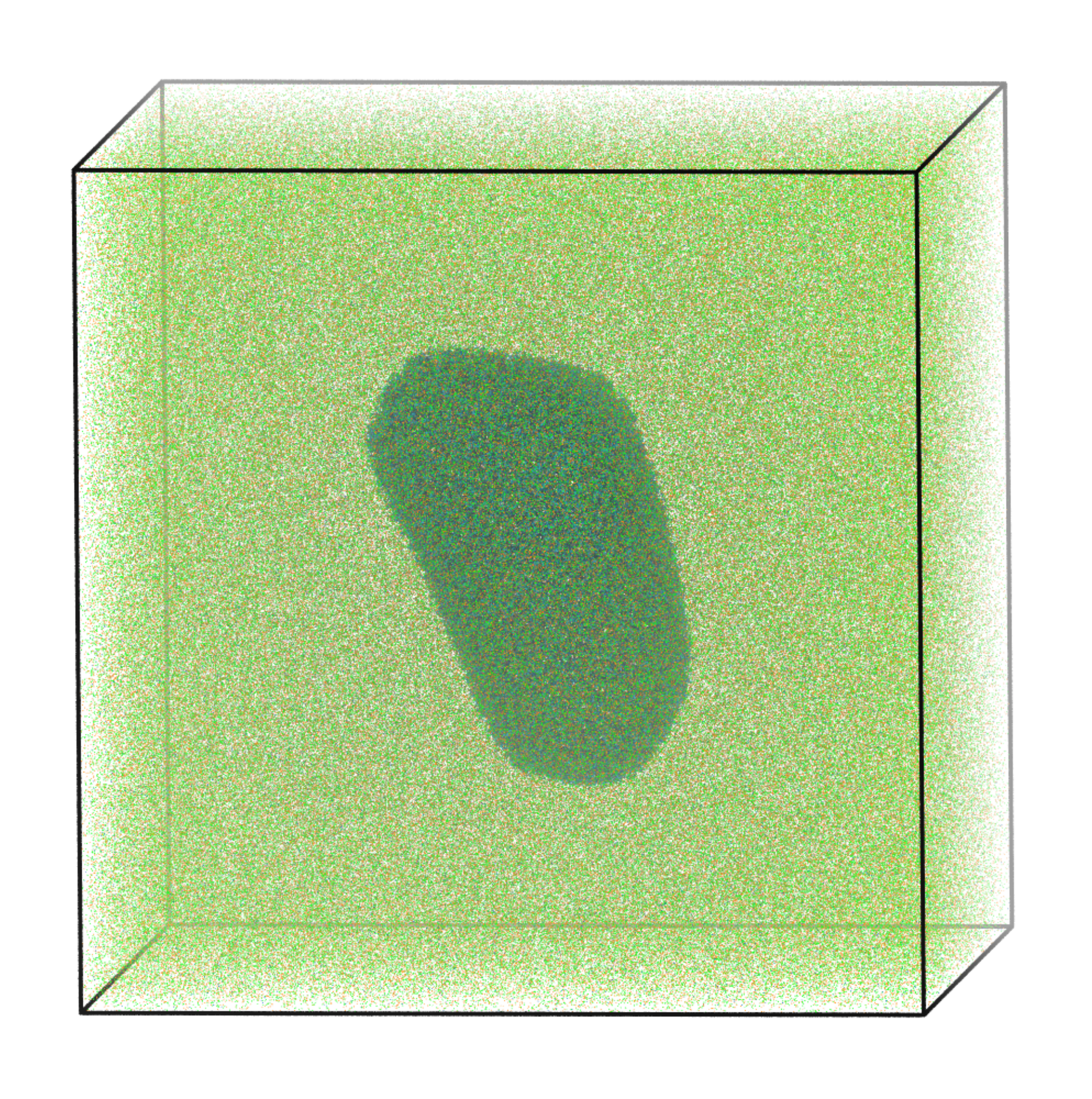
In the coming years, we plan to continue to extend and refine our ion models, which will account for additional ion species and classes of ionic interactions. To show the potential for these approaches, we also performed proof-of-principle simulations of the HIV-1 capsid with explicit ions (right), where we were able to describe the dynamics of ions around the capsid shell.
